
Polymerase chain reaction (PCR)
Polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it to a large enough amount to study in detail. PCR was invented in 1984 by the American biochemist Kary Mullis at Cetus Corporation. It is fundamental to much of genetic testing including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory and clinical laboratory research for a broad variety of applications including biomedical research and criminal forensics.[14] [14a]
PCR amplifies a specific region of a DNA strand (the DNA target). Most PCR methods amplify DNA fragments of between 0.1 and 10 kilo base pairs (kbp) in length, although some techniques allow for amplification of fragments up to 40 kbp.[14b]
Principles
A basic PCR set-up requires several components and reagents [14c]:
- DNA template that contains the DNA target region to amplify;
- DNA polymerase; an enzyme that polymerizes new DNA strands; heat-resistant Taq polymerase is especially common, [14d] as it is more likely to remain intact during the high-temperature DNA denaturation process;
- two DNA primers that are complementary to the 3' (three prime) ends of each of the sense and anti-sense strands of the DNA target (DNA polymerase can only bind to and elongate from a double-stranded region of DNA; without primers, there is no double-stranded initiation site at which the polymerase can bind); [14e] specific primers that are complementary to the DNA target region are selected beforehand, and are often custom-made in a laboratory or purchased from commercial biochemical suppliers;
- deoxynucleoside triphosphates, or dNTPs (sometimes called "deoxynucleotide triphosphates"; nucleotides containing triphosphate groups), the building blocks from which the DNA polymerase synthesizes a new DNA strand;
- a buffer solution providing a suitable chemical environment for optimum activity and stability of the DNA polymerase;
- bivalent cations, typically magnesium (Mg) or manganese (Mn) ions; Mg2+ is the most common, but Mn2+ can be used for PCR-mediated DNA mutagenesis, as a higher Mn2+ concentration increases the error rate during DNA synthesis; [14f] and monovalent cations, typically potassium (K) ions
Procedure
Typically, PCR consists of a series of 20-40 repeated temperature changes, called thermal cycles, with each cycle commonly consisting of two or three discrete temperature steps . The cycling is often preceded by a single temperature step at a very high temperature (>90°C (194°F)), and followed by one hold at the end for final product extension or brief storage.
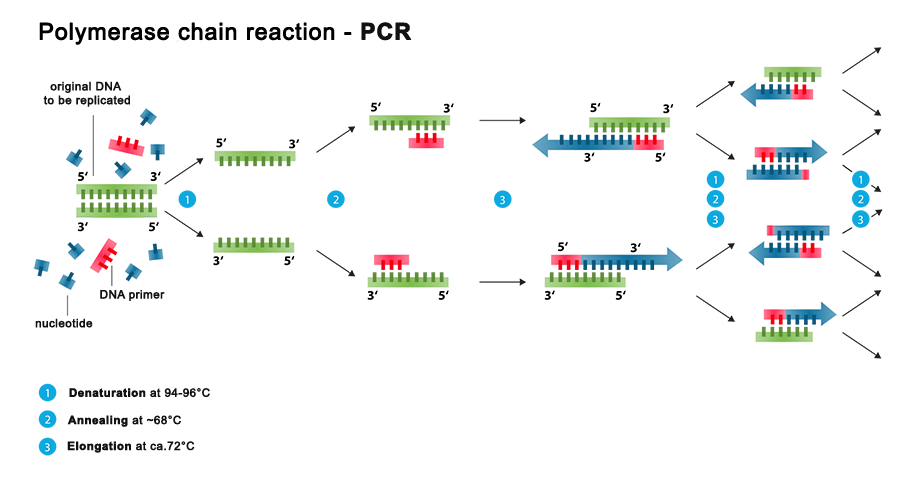
Schematic representation of PCR process [14]
Image source: Wikimedia
Steps common to most PCR methods
Denaturation: This step is the first regular cycling event and consists of heating the reaction chamber to 94-98°C (201-208°F) for 20–30 seconds. This causes DNA melting, or denaturation, of the double-stranded DNA template by breaking the hydrogen bonds between complementary bases, yielding two single-stranded DNA molecules.

Annealing: the reaction temperature is lowered to 50–65 °C (122–149°F) for 20–40 seconds, allowing annealing of the primers to each of the single-stranded DNA templates. Two different primers are typically included in the reaction mixture: one for each of the two single-stranded complements containing the target region. The primers are single-stranded sequences themselves, but are much shorter than the length of the target region. The temperature must be low enough to allow for hybridization of the primer to the strand, but high enough for the hybridization to be specific.
Extension: The temperature at this step depends on the DNA polymerase used. The optimum activity temperature for the thermostable DNA polymerase of Taq polymerase is approximately 75-80-°C (167-176°F), [14g] [14h] though a temperature of 72°C (162°F) is commonly used with this enzyme. In this step, the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding free dNTPs from the reaction mixture that is complementary to the template in the 5'-to-3' direction, condensing the 5'-phosphate group of the dNTPs with the 3'-hydroxy group at the end of the nascent (elongating) DNA strand.
The processes of denaturation, annealing and elongation constitute a single cycle. Multiple cycles are required to amplify the DNA target to millions of copies. To check whether the PCR successfully generated the anticipated DNA target region (also sometimes referred to as the amplimer or amplicon), agarose gel electrophoresis may be employed for size separation of the PCR products. The size of the PCR products is determined by comparison with a DNA ladder, a molecular weight marker which contains DNA fragments of known sizes, which runs on the gel alongside the PCR products.
© www.humankaryotype.com