
Fluorescent in situ hybridization (FISH)
Fluorescent in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only those parts of a nucleic acid sequence with a high degree of sequence complementarity. In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence of interest. FISH was developed by biomedical researchers in the early 1980s [10] to detect and localize the presence or absence of specific DNA sequences on chromosomes.
First, a probe is constructed. The probe must be large enough to hybridize specifically with its target but not so large as to impede the hybridization process. The probe is tagged directly with fluorophores, with targets for antibodies or with biotin. Tagging can be done in various ways, such as nick translation, or Polymerase chain reaction using tagged nucleotides. Then, an interphase or metaphase chromosome preparation is produced. The chromosomes are firmly attached to a substrate, usually glass. Repetitive DNA sequences must be blocked by adding short fragments of DNA to the sample. The probe is then applied to the chromosome DNA and incubated for approximately 12 hours while hybridizing. Several wash steps remove all unhybridized or partially hybridized probes. The results are then visualized and quantified using a microscope that is capable of exciting the dye and recording images. [10]
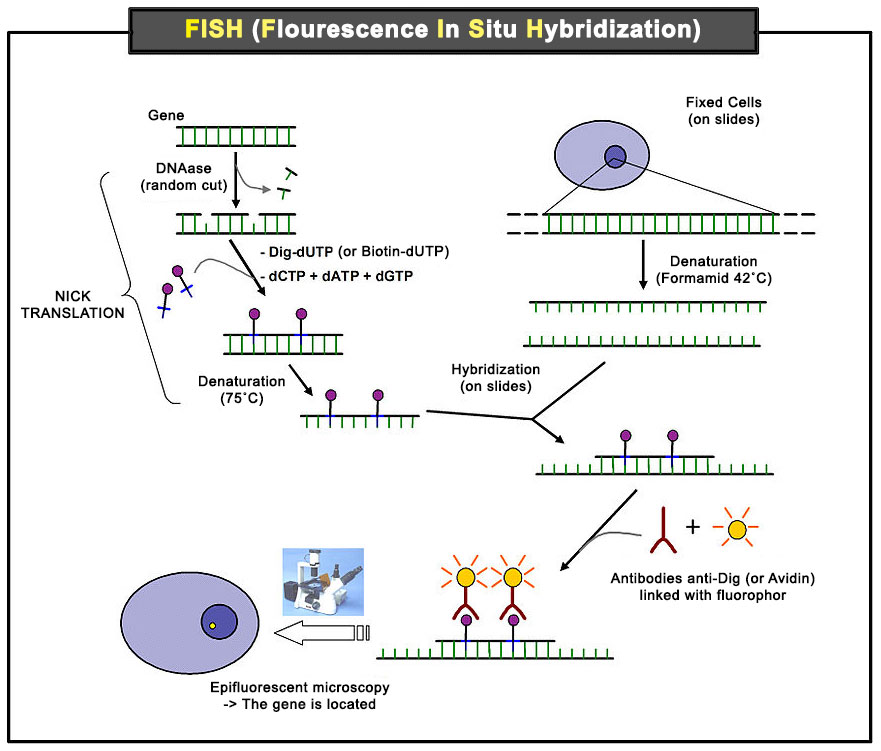
Scheme of the principle of the FISH Experiment to localize a gene in the nucleus. [10]
Legend: Nick translation [11] (or head translation), developed in 1977 by Peter Rigby and Paul Berg, is a tagging technique in molecular biology in which DNA Polymerase I is used to replace some of the nucleotides of a DNA sequence with their labeled analogues, creating a tagged DNA sequence which can be used as a probe in fluorescent in situ hybridization.
Image source: Wikimedia
Molecular hybridization between complementary DNA sequences has been used to develop a new technology allowing the visualization of biological material at a scale that lies between the microscopic and the molecular scale, inaccessible to microscopic visualization. Each cellular or sub-cellular structure visible with the microscope and containing a nucleic acid molecule (chromosome, nucleus or ribosome) can be treated in situ after fixation on a slide that makes the nucleic acid molecule (RNA or DNA) accessible for a specific fluorochrome-carrying probe. The fixation of the fluorescence shows that the specific sequence is present and allows its localization. The absence of fluorescence shows that the sequence is absent. FISH brings an important complementary technique to the cytogenetic or genetic analysis because it gives information on the chromosome localization of a sequence, whether it has been deleted or translocated. [6]
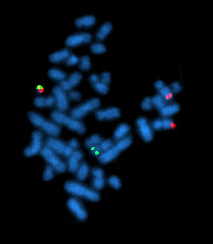
A metaphase cell positive for the bcr / abl rearrangement, translocation 9 - 22 (associated with chronic myelogenous leukemia) using FISH. The chromosomes can be seen in blue. The chromosome that is labeled with green and red spots (upper left) is the one where the rearrangement is present. [10]
Image source: Wikimedia
Some karyotypic anomalies, not accessible to conventional cytogenetics (microdeletions, uniparental disomies) can be detected or identified by techniques such as fluorescent in situ hybridization (FISH) combining karyotyping and molecular biology. [6]
© www.humankaryotype.com